
INSPIRE
Our inclusive postdoctoral fellowship combines cutting edge research with cutting edge teaching
to train the next generation of scientists/educators.
Interested in Becoming an INSPIRE Fellow?
We are currently recruiting fellows to join us for the 2025-2026 academic year.
About the Program
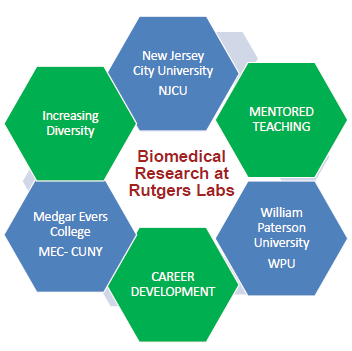
The INSPIRE (IRACDA New Jersey/New York for Science Partnerships in Research and Education) Postdoctoral Research and Education Program at Robert Wood Johnson Medical School provides up to three years of mentored research experience in biomedical fields at the medical school or Rutgers University. The postdoctoral fellowship program also provides training in educational methods, including mentored teaching at a nearby partner institution, and career development. INSPIRE is sponsored by NIH-IRACDA (Institutional Research and Career Development Award).
Two Important Program Goals
- Prepare university-trained PhD scholars for successful careers as scientist-educators
- Increase the participation of undergraduate populations in biomedical science research fields
The program has also increased interaction of medical school faculty with students and faculty from our partner universities and colleges in New York and New Jersey. It has enhanced the professional development of other trainees at Rutgers who participate in INSPIRE activities (e.g. workshops on scientific writing, courses on scientific teaching).
Our rigorous training in basic research typically generates an independent research program which INSPIRE Fellows can use to launch their own labs and high-impact publications.
70% of INSPIRE Fellows obtain tenure-track academic jobs.
INSPIRE Training
- 70% time for high-powered research, achieving high-impact publications
- 30% time for career skills, including mentored teaching at one of our partner institutions
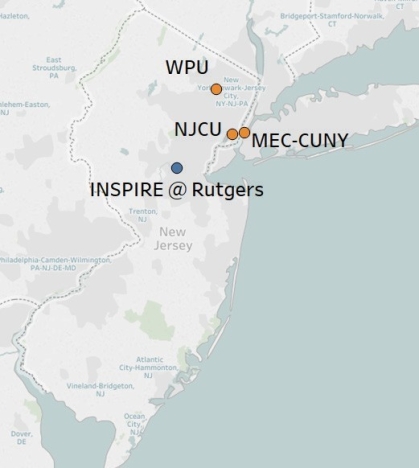
Partnering on the INSPIRE program are three nearby primarily undergraduate-serving institutions:
Alumni Testimonials
Watch short video testimonies from some of our successful alumni.
Contact Us
Leadership Team

INSPIRE Co-Director Martha Soto is PI of the INSPIRE grant and professor of the Department of Pathology. She was director of the Postdoctoral Career Development Program from 2010 to 2017, when in 2017 she helped Rutgers launch the campus-wide Office of Postdoctoral Advancement. She is also the founder and faculty advisor for the BIO Links (Biosciences Links to Teaching) K-12 Outreach Program. In Bio Links, graduate students and postdoctoral fellows at RWJMS and Rutgers partner with local middle school and high school science teachers, serving diverse student populations.
INSPIRE Co-Director Gary A. Brewer is co-PI of INSPIRE and professor of the Department of Biochemistry and Molecular Biology. He is a mentor and writing coach for the National Research Mentoring Network (NRMN).
INSPIRE Co-Director Detlev Boison is the newest member of the team. He is a professor in the Department of Neurosurgery at RWJMS.
Patricia Irizarry-Barreto is program support coordinator of INSPIRE and research lab Manager in the Department of Pathology. She has a background in molecular biology in mRNA degradation. As an NSF-GK-12 fellow she developed skills in science communication and student development with expertise in Higher Education outreach programs. She has been recognized for Innovation in Teaching and Learning and Team Work in 2016 and 2020.
Partner School Program Coordinators
- Emily Monroe, Assistant Professor at William Paterson University
- Cindy Arrigo, Associate Professor at New Jersey City University
- Edward J. Catapane, Professor at Medgar Evers College-CUNY
Faculty and program coordinators from these schools play an integral role in the educational training and professional development of our INSPIRE Fellows.
Location
The INSPIRE program is located in the Research Tower on Piscataway Campus. Most of our events happen here. However, our fellows conduct their daily research in various labs across the Rutgers campuses in New Brunswick, Piscataway, and Newark.
INSPIRE Postdoctoral Research and Education Program
Robert Wood Johnson Medical School
Research Tower
675 Hoes Lane West
Piscataway, NJ 08854
Application Details
Why Apply?
- One of the most diverse academic communities in the nation is ready to welcome you
- Competitive funding for up to three years
- Dedicated, strong and supportive community
- Your career development is our priority:
- 75% mentored research at a Rutgers laboratory
- 25% mentored teaching at one of our partner institutions
We encourage applications from people underrepresented in science.
Especially if you are preparing for a U.S. academic faculty position, if you are passionate about research, and care about teaching, come join us!
Application Process
Phase 1 of the application involves writing the equivalent of a one-page grant proposal. Please identify your (potential) research mentor on the proposal, and work with them to develop a strong research proposal. If you need our advice finding the right research lab for your interests, please reach out to us.
Once you have submitted your application, you will receive a link via email to continue on to phase 2.
Start Phase 1 of the Application Process
Eligibility and Pertinent Information
- Basic research in biomedical fields, broadly defined. Our fellows are typically in biochemistry, biophysics, cancer biology, cell/developmental biology, computational/quantitative biology or bioinformatics, genetics/genomics, immunobiology, microbiology, molecular biology, neuroscience
- U.S. citizen or Permanent Resident only (per NIH requirements)
- Postdoctoral fellows with 0-1 years of postdoctoral training will receive preference
Important Dates and Deadlines
- Review of applications usually begins on June 1. This year we are currently open for applications now (Fall 2025).
- Full-day interviews begin soon.
- Fellowships begin in September 1st or depending on your appointment date.
- For a September start date the deadline is May 26, but due to rolling admissions, applications are still welcome over the summer and fall. This year we are running a fall recruitment with a later start date.
- Late admissions may be accommodated by the program
- Please identify your (potential) research mentor on the one page proposal, and work together to develop a strong research proposal
INSPIRE supports equal opportunity in employment for all qualified persons and does not discriminate on the basis of race, color, religion, gender, sexual orientation, social class, national origin, or disability unrelated to job requirements.
Questions?
Program Support Coordinator Patty Irizarry can address any other questions you have about your application.
Frequently Asked Questions
Accordion Content
-
INSPIRE Fellows are mentored by faculty who have strong records of funding, mentoring, and publishing.
Our Fellows receive mentoring and support for their teaching at our partner minority-serving institutions in New Jersey and New York. INSPIRE fellows are diverse themselves.
INSPIRE has had outstanding success placing Fellows in tenure-track faculty positions (70%).
-
Applicants must be U.S. citizens or permanent residents, and ideally with no more that 0-3 years of prior postdoctoral experience. See full eligibility details above.
-
We like to see evidence that you care about teaching. We do not expect extensive teaching experience since the program will provide this.
-
No, we are open to all applicants, including members of underrepresented groups. We particularly look for individuals who have shown an interest in improving science education for all. We very much welcome Fellows from all groups underrepresented in STEM, including disabled students.
-
This is first and foremost a research program, so it matters which lab you plan to join. Especially since our application involves writing the equivalent of a short (one page) grant. If you need our advice finding the right research lab for your interests, please reach out to us.
-
Fellows can work with research mentors at Rutgers who do biomedical research. If you are not sure, please check with us and we are happy to point you in the right direction.
-
All applicants give a five-minute presentation with two slides that introduce and show the main findings of their thesis. This is directly followed by a five-minute talk with one slide to introduce the proposed postdoc project. Applicants present to the INSPIRE Leadership, Admissions Committee, and selected current Fellows.
-
Yes, all postdocs with 0-3 years of experience and are U.S. citizens or permanent residents are eligible for the program.
-
We meet at least once a month with the larger Fellows group and support additional smaller groups such as writing groups.
-
We pay based on the NIH Postdoctoral pay scale.
-
We pay the healthcare costs for the Fellow, and we give each Fellow travel and supply funds.
-
Year 1: Participation in a comprehensive Scientific Teaching Workshop in the spring.
Year 2: Suggested minimum of 8 hours of independent in-class teaching with supervision and feedback by the Teaching Mentor.
Year 3: Suggested 16 hours of independent in-class teaching with supervision and feedback by the Teaching Mentor.
Faculty Mentors

Applicants are encouraged to contact faculty members leading a Robert Wood Johnson Medical School or Rutgers biomedical research lab, depending on their research interest. You can search for a mentor in the list below, but you are not limited to these.
Our fellows' research mentors are faculty members from across the Rutgers campuses in New Brunswick, Piscataway, and Newark. All Rutgers labs are eligible as long as the research will be conducted in the biomedical fields (broadly defined).
We welcome inquiries from Rutgers and Rutgers Health faculty who are interested in joining the INSPIRE program. If you are committed to mentoring postdoctoral fellows who want to obtain academic jobs, and you have an active research lab that could incorporate a talented postdoc who can be funded by INSPIRE for up to three years, we would love to hear from you. You can start by recruiting a top candidate in your field from around the country to your lab when you attend conferences for example. Once you have a candidate, you can ask them to apply for INSPIRE funding.
Find a Mentor
Accordion Content
-
Name Area of Expertise Research Interest with links Abraira, Victoria, PhD Neuroscience Fundamental principles of sensory neurobiology; spinal cord circuits
Anthony, Tracy, PhD Nutritional Science Amino Acid Control of Protein Synthesis and Gene Expression
Aston-Jones, Gary, PhD Brain Health Institute Psychiatry Brain Mechanisms of Motivation and Cognitive Processes Cognition and Addiction.
https://brainhealthinstitute.rutgers.edu/faculty/core-investigators/gary-aston-jones-lab/
Barr, Maureen, PhD Genetics Generation of sexual behaviors and the molecular basis of human genetic diseases of cilia.
Boison, Detlev, PhD Neurosurgery To translate fundamental mechanisms of biochemistry and energy metabolism into novel therapeutic approaches for the treatment of neurological conditions.
https://molbiosci.rutgers.edu/faculty-research/faculty/faculty-detail/79-a-b/720-boison-detlev
Brewer, Gary, PhD Biochemistry and Molecular Biology Regulation of Gene Expression; Genomics
https://molbiosci.rutgers.edu/faculty-research/faculty/faculty-detail/79-a-b/125-gary-brewer
Copeland, Paul, PhD Biochemistry & Molecular Biology Regulation of Gene Expression at the Translational Level, Incorporation and Utilization of Selenocysteine.
https://molbiosci.rutgers.edu/faculty-research/faculty/faculty-detail/80-c-d/134-paul-r-copeland
DiCicco-Bloom, Emanuel, MD Neuroscience and Cell Biology Pediatrics Regulation of Developmental and Adult Neurogenesis, Cell Cycle Mechanisms.
Dreyfus, Cheryl, PhD Neuroscience and Cell Biology The Role of the Trophic Factors and Glial Cells.
Driscoll, Monica, PhD Molecular Biology and Biochemistry Developmental Neurogenetics, Molecular Genetics of Neuronal Cell Death, Mechanosensory Transduction in Touch and Feeling, Molecular Mechanisms of Aging
https://sites.rutgers.edu/driscoll-lab/people/dr-monica-driscoll/
Duffy, Siobain, PhD Ecology, Evolution & Natural Resources Emerging Viruses, Experimental Evolution, Bioinformatics, Bacteriophage.
Ellison, Christopher, PhD Genetics Computational Biology, Genomics, Molecular Genetics.
Fan, Huizhou, PhD Pharmacology Pathogenesis and Intervention of Sexually Transmitted Infections.
https://molbiosci.rutgers.edu/faculty-research/faculty/faculty-detail/81-e-f/142-huizhou-fan
Freundlich, Joel , PhD Pharmacology, Physiology & Neuroscience Computation, chemistry, and biology to develop new tools that contribute to our understanding of the pathogenesis of infectious diseases.
Gao, Nan, PhD Biological Sciences Human commensal bacteria, pathogen, and intestinal inflammation.
Irvine, Kenneth, PhD Molecular Biology and Biochemistry Cell-Signaling, Pattern Formation, Growth Control, Developmental Glycobiology.
Jacinto, Estela, PhD Biochemistry and Molecular Biology Signal Transduction, Cancer, Insulin, Metabolism, T Lymphocytes.
Kiledjian, Mike, PhD Cell Biology and Neuroscience RNA-Protein Interactions in the Regulation of Mammalian Mrna Turnover. RNA-Binding Proteins in Human Genetics Disorders
McKim, Kim, PhD Genetics Double Strand Break Repair, Genetic Recombination, Chromosome Pairing, Chromosome Segregation, Kinesin Motor Proteins.
Millonig, James, PhD Graduate School of Biomedical Sciences Neuroscience & Cell Biology DHuman Neurodevelopmental Disorders, Autism.
Monahan, Kevin, PhD Molecular Biology & Biochemistry Gene regulatory mechanisms that specify diverse cell-types in the mammalian nervous system and maintain their identity for the life of an organism.
Nakamura, Tetsuya, PhD Evolutionary and Developmental biology The genetic mechanisms underlying the fish diversity and fish-to-tetrapod transition.
Nanda, Vikas, PhD Biochemistry Protein Evolution and Folding, Computational De Novo Design of Proteins and Biomimetics.
Omary, Bishr PhD Biomedical Sciences Keratin protein aggregates
Quadro, Loredana, PhD Food Science Nutritional Biochemistry.
Roepke, Troy, PhD Animal Sciences Environmental influences and stresses on the physiological functions of organisms.
Routh, Vanessa, PhD Pharmacology, Physiology & Neuroscience Role Of Hypothalamic Glucose Sensing Neurons In Glucose And Energy Homeostasis.
https://njms-web.njms.rutgers.edu/profile/myProfile.php?mbmid=routhvh
Runnels, Loren, PhD Pharmacology Developmental Biology, Magnesium and Calcium Homeostasis and Signaling, Brain Injury.
https://molbiosci.rutgers.edu/faculty-research/faculty/faculty-detail/87-q-r/115-loren-runnels
Samuels, Benjami, PhD Psychology Chronic Stress Paradigms For Both Sexes.
Schindler, Karen, PhD Genetics Signal Transduction, Meiosis, Aneuploidy, Gamete Formation and Reproduction.
Singson, Andrew, PhD Genetics Reproductive Biology and Cell-Cell Interactions in C. Elegans.
Soto, Martha, PhD Pathology C. elegans model for Cell Polarity and Cell Migrations During Development and Disease.
https://rwjms.rutgers.edu/department/pathology-laboratory-medicine/soto-lab
Stock, Ann, PhD Biochemistry & Molecular Biology Structure/Function Analysis of Signal Transduction Proteins.
Verzi, Michael, PhD Genetics Basic intestinal development and homeostasis, as well as various aspects of intestinal health.
Neurology Department https://ifh.rutgers.edu/faculty_staff/auriel-a-willette-ba-ms-phd/ Zhang, Huaye, PhD Neuroscience and Cell Biology Regulation of Dendritic Spine Morphogenesis and Synaptic Plasticity.
https://molbiosci.rutgers.edu/faculty-research/faculty/faculty-detail/91-y-z/210-huaye-zhang
Current Fellows

Edward Chuang
Department of Molecular Biology and Biochemistry
Email: edward.chuang@rutgers.edu
Research Topic: Trafficking of misfolded and aggregated proteins

Joshua Danoff
Department of Molecular Biology and Biochemistry
Email: joshua.danoff@rutgers.edu
Research Topic: Chromatin-mediated adaptations to environmental context in sensory neurons

Caroline Shwaner
Genetics Department
Email: caroline.schwaner@rutgers.edu
Research Topic: The genetic mechanisms underlying digit evolution in tetrapods

Katherine Denney
Psychology
Email: kd945@scarletmail.rutgers.edu
Research Topic: Sex differences in mood disorders

Katherine Jacobs
Department of Genetics/HGINJ
Email: kj490@rwjms.rutgers.edu
Research Topics: I study relationships between extracellular matrix, sensory cilia and extracellular vesicles with the aim of understanding how ECM impacts ciliary function.

Wesley Evans
Department of Cell Biology and Neuroscience
Email: we41@dls.rutgers.edu
Research Topic: Astrocyte physiology in the striatum in parkinsonianism.

Katherine Maniates
Waksman Institute of Microbiology
Email: maniates@waksman.rutgers.edu
Research Topic: Identification of genes and molecular mechanisms for fertilization in C. elegans

Shams Shams Department of Chemical Biology
Email: ss4459@rwjms.rutgers.edu
Research Topic: Role of Aminoacid metabolism and Nutrition in Cancer

Karen Olson
Department of Biological Sciences
Email: olsonvkaren@gmail.com
Research Topic: Population genetics, bat and cave microbiomes

Benton Purnell
Department of Neurosurgery
Email: benton-purnell@uiowa.edu
Research Topic: Role of adenosine in sudden unexpected death in epilepsy (SUDEP)

Timothy J. Stanek
Human Genetics Institute of New Jersey
Email: timothy.stanek@rutgers.edu
Research Topic: Role of R-loops in regulating chromatin architecture

Gwyndolin Vail
Pharmacology, Physiology, and Neuroscience
Email: gvail@gsbs.rutgers.edu
Research Topic: Sex differences in central sensing and regulation of glucose during exercise

Marcus Begley
Genetics, HGINJ
Email: mb2389@hginj.rutgers.edu
Research Topic: Meiotic spindle biophy
INSPIRE Alumni

Accordion Content
-
Melanie Johnston
Tenure-track Assistant Professor of Biology
Northern Arizona University, AZAlex Cope (2021-2024)
Postdoctoral Associate - Rokas lab
Vanderbilt University, TNDarlingtina Esiaka (2021-2023)
Tenure-track Assistant Professor
University of KY -College of Medicine.Kelly Kyker-Snowman (2019-2023)
Assistant Teaching Professor of Biomedical Engineering
Rutgers University, NJMaria Agapito (2013-2015)
Tenure-track Assistant Professor of Biology
Bard College, NJJorge Avila (2020-2022)
Assistant Director, Undergraduate Research Center-Science
University of California Los Angeles, CADenver Baptiste (2019-2020)
Tenure-track Assistant Professor
St. Peter's University, NJSofya Borinskaya (2015-2019)
Assistant Professor
Saint Elizabeth University, NJGerialisa V. G. Case (2019-2020)
In transitionVicky DiBona (2017-2021)
Tenure-track Assistant Professor
Saint Anselm College, NHKeith Feigenson (2011-2014)
Tenure-track Assistant Professor, Dept. of Psychology
Albright College, PAJessica Fellmeth (2018-2021)
Tenure-track Assistant Professor
Millersville University, PABenjamin Griffel (2011-2014)
Tenure-track Professor of Biology and Mathematics
Bard High School Early CollegeEric Ho (2010-2013)
Tenured Associate Professor, Dept. of Biology with affiliation in Computer Science Department
Lafayette College, PAAmber Krauchunas (2013-2016)
Tenure-track Professor
University of DelawareJason Lunden (2013-2016)
Investigator
Hussman Institute for AutismMatthew Marcello (2010-2013)
Tenured Associate Professor, Dept. of Biology and Health Sciences
Pace University, NYKenneth McGuiness (2019-2021)
Assistant Professor
Caldwell University, NJHarita Menon (2014-2017)
Adjunct faculty
Georgian Court University, NJDina Navon (2020-2022)
Assistant Professor, Biology Department
University of the Fraser Valley, British Columbia, CanadaMichael Nestor (2012-2014)
Investigator
Hussman Institute for Autism
Adjunct Faculty
University of Maryland, MDKenny Nguyen (2012-2015)
Head of Computational Science & Engineering
Children's Hospital of Philadelphia (CHOP), PAInna Nikonorova (Associate Fellow, 2015-2017)
Postdoctoral Fellow
Rutgers University, NJOyenike Olabisi (2010-2012)
Tenured Associate Professor, Dept. of Biological Sciences
University of DelawareAshley Pettit (2014-2017)
Faculty, Dept. of Biological Sciences
Seton Hall University, NJSuzanne Quartuccio (2014-2017)
Tenure-track Assistant Professor
Seton Hall University, NJMatthew Roche (2012-2015)
Assistant Professor of Psychology
New Jersey City University (on leave), NJJessica Saalfield (2018-2020)
Tenure-track Assistant Professor
Penn State University, PAStephen Shannon (2012-2016)
Medical Writer
ETHOS Health CommunicationsPragati Sharma (2017-2019)
HealthAdvance Manager
Rutgers Office of Research and Economic Development, NJAlthea Stillman (2011-2014)
Associate Director of Life Sciences
IP Group plcDavid Swope (2012-2015)
Tenure-track Assistant Professor, Cooper Medical School
Rowan University, NJKrishna Tobon (2012-2014)
Adjunct Faculty
Fairleigh Dickinson University, NJAndre Wallace (2011-2014)
Tenure-track Assistant Professor of Biological Sciences
Fairleigh Dickinson University, NJJonathon Walsh (2018-2021)
Genome engineer
Colossal Biosciences, TX
Other Resources
Accordion Content
-
ASCB (American Society for Cell Biology): Annual Conference by the MAC (Minority Affairs Committee)
SACNAS (Society for Advancement of Chicanos/Hispanics and Native Americans in Science)
MACUB Conference (Metropolitan Association of College & University Biologists): A regional conference for undergraduate researchers, many who are underrepresented.
Keystone Symposia in Biomedical and Life Sciences
The National Center for Faculty Development and Diversity: An independent professional development, training, and mentoring community of over 83,000 graduate students, post-docs, and faculty members.
Resources for Postdocs
Postdoc Resources
Find support for postdoctoral fellows including writing assistance, career development, and more.
News & Events
News
-
- Thank you Dr. Marc Muñiz for talking to our fellows about "How to write good exam questions".
- Congratulations Dr. Wesley Evans for publishing: https://onlinelibrary.wiley.com/doi/epdf/10.1002/glia.24679
- INSPIRE Research and Teaching Seminar will be on Friday May 2nd, 2025. Our keynote speaker will be: Dr. Angeline Duke, Assistant Professor in the Department of Neuroscience at the University of Minnesota.
-
- INSPIRE Writing Groups meeting on Dec 5th
- INSPIRE Cafe on Nov 21 Guest Speaker: Dr. Julia Schmitz from Piedmont University.
- INSPIRE Retreat on October 17 Key note: Dr. Jessica Fellmeth, “Genes, Pedagogy, and Persistence: My Evolution to Assistant Professor". Afternoon program at the Rutgers Gardens.
- INSPIRE Fall Writing Groups started on October 2024.
- Teaching Seminar by Dr. Ramazan Gungor: Designing and Delivering Effective Distance Education in Higher Education.
- INSPIRE Fellows, INSPIRE Leadership from Rutgers and our Partner Institutions attended the IRACDA Conference 2024 at UNC- Chapel Hill in North Carolina.
- INSPIRE hosted a Scientific Teaching Seminar, "Designer Jackets and Synthetic Threads for Probing the Immune Response" on May 10, 2024, featuring a keynote lecture by Catherine Grimes, PhD, a Professor of Chemistry and Biochemistry at the University of Delaware.
-
- INSPIRE hosted our annual 2023 Scientific Teaching Seminar on April 25, with guest speaker Dr. Erich Jarvis.
- Dr. Patty Irizarry joined INSPIRE in February. Dr. Irizarry is a Molecular Biologist and Outreach Professional who is currently serving as our new Program Support Coordinator.
- We hosted the 2023 INSPIRE Retreat on Nov. 3, with keynote speaker Dr. Maria Agapito.
-
- INSPIRE hosted our 2022 Annual Scientific Teaching Seminar on May 17, with invited speaker Dr. Robin Wright.
- We hosted another visit from our Advisory Board Members on April 11-12. Our program benefited greatly from their visit in 2019, with their expert advice and report.
- INSPIRE funded five undergrads from our partner schools to attend the 2022 RISE at Rutgers summer research program, mentored by INSPIRE fellows in their labs.
Generous funding from the office of Rutgers' previous SVPAA Dr. Barbara Lee, and now EVPAA (Executive Vice President for Academic Affairs) Dr. Prabhas V. Moghe, Rutgers' Chief Academic Advisor, helped to make this great opportunity possible.
We have successfully run this RISE-INSPIRE program for 5 years, since the renewal of INSPIRE grant in 2017. This RISE-INSPIRE Summer Research program also helps support the STEM diversity pipeline at Rutgers. As RISE-INSPIRE Scholars, participants are also eligible for the Summer Undergraduate Pipeline to Excellence at Rutgers Graduate (SUPER Grad) fellowship program if they return to Rutgers for graduate study.
-
- INSPIRE hosted our 2021 Annual Fall Seminar Series virtually on Oct. 28.
- INSPIRE co-hosted the NIH-IRACDA 2021 National Conference, virtually, for 3 days on June 28-30. Participants also shared updates on Twitter at @iracda2021.
- Congratulations to two of our fellows who exited the program for academic roles:
- Vicky DiBona
Tenure-track Assistant Professor
Saint Anselm College, NH - Jessica Fellmeth
Tenure-track Assistant Professor
Penn State Schuykill, PA
- Vicky DiBona
-
- 2020 Annual Scientific Teaching Seminar, Dec. 9: “Towards More Inclusive Active Learning Classrooms: How groups of students are differentially impacted by active learning," presented by Dr. Sara Brownell, Associate Professor, Arizona State University.
- 2020 INSPIRE Annual Retreat, Oct. 16, including a Morning Workshop from NCFDD, "Building a Publishing Pipeline: Concrete Strategies for Increasing Your Writing Productivity," led by Professor Erin Furtak, University of Colorado Boulder, Member of the National Center for Faculty Development & Diversity (NCFDD).
- 2019 INSPIRE Fall Retreat, with INSPIRE Alumni Dr. Matthew R. Marcello, Associate Professor at Pace University, who gave an exciting speech and shared more advice with INSPIRE fellows at our luncheon afterwards.
- 2019 Rutgers Active Learning Symposium, May 22 at the Richard Weeks Hall of Engineering on Busch Campus in Piscataway, NJ.
- 2019 INSPIRE Annual Scientific Teaching Seminar, May 9, with Dr. Marybeth Gasman from University of Pennsylvania, who joined Rutgers in fall 2019.
- 2019 NIH-IRACDA conference at the University of Michigan, Ann Arbor, with presentations by our INSPIRE fellows.
- 2018 INSPIRE Inaugural Fall Retreat, with two featured speakers, Eric Ho and John Kerrigan.
- 2018 NIH-IRACDA conference at the University of Georgia, with presentations by our INSPIRE fellows.
- 2018 INSPIRE Annual Scientific Teaching Seminar, with Dr. Scott Freeman from University of Washington and INSPIRE alumnus Dr. Jason Lunden, who was featured on the journal Nature in 2015: "A researcher who studies autism-like behaviour in mice takes inspiration from his own condition."
"Jason Lunden was diagnosed with Asperger's syndrome while doing a PhD in neuroscience. Now a postdoc at the Rutgers Robert Wood Johnson Medical School in New Brunswick, New Jersey, he conducts research on stress in mice that exhibit autism-like behaviours." -
2017 NIH-IRACDA conference, June 4-6, at the University of Alabama at Birmingham with presentations by three of our INSPIRE fellows.
-
2015 INSPIRE Annual Scientific Teaching Seminar with a presentation by Dr. April Hill from University of Richmond.


